OpenHexa, Bluesquare’s new Data Integration Platform

We often hear about the scarcity of health data coming from low resource countries. However, in the last decade, we at Bluesquare have experienced a significant increase in the volume of data collected through the expansion of software solutions such as DHIS2.
With the development of data transmission through multiple communication networks, it can become hard to keep track of all the data that public facilities, private actors, NGOs running campaigns, research projects or logistics systems collect. It is even harder to mobilize these various sources of data to create meaningful analysis and reports. Yet, these can help guide public health decisions.
To help solve this problem, Bluesquare is proud to introduce OpenHexa, our open-source data integration platform targeted at health data professionals.
Solving the key challenges of data processing : Exploration, Extraction, Visualisation
OpenHexa focuses on four key capabilities:
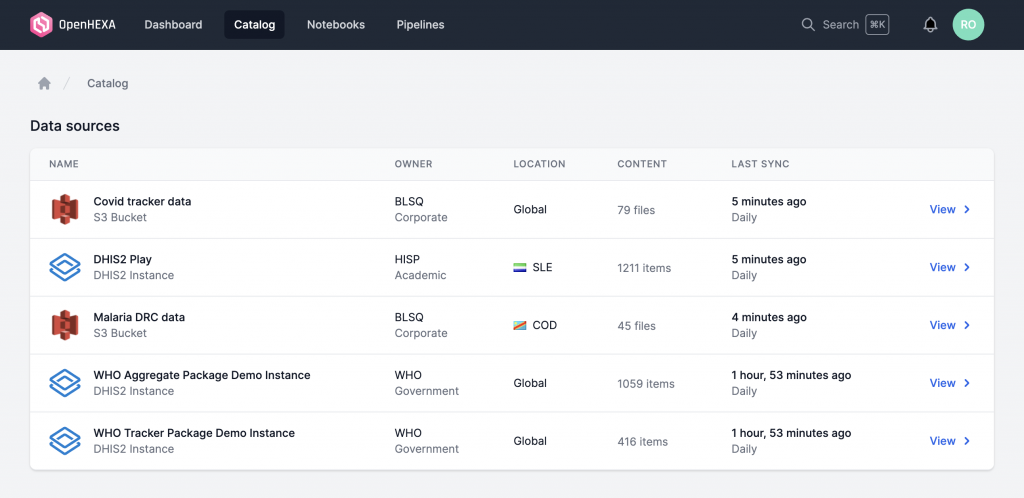
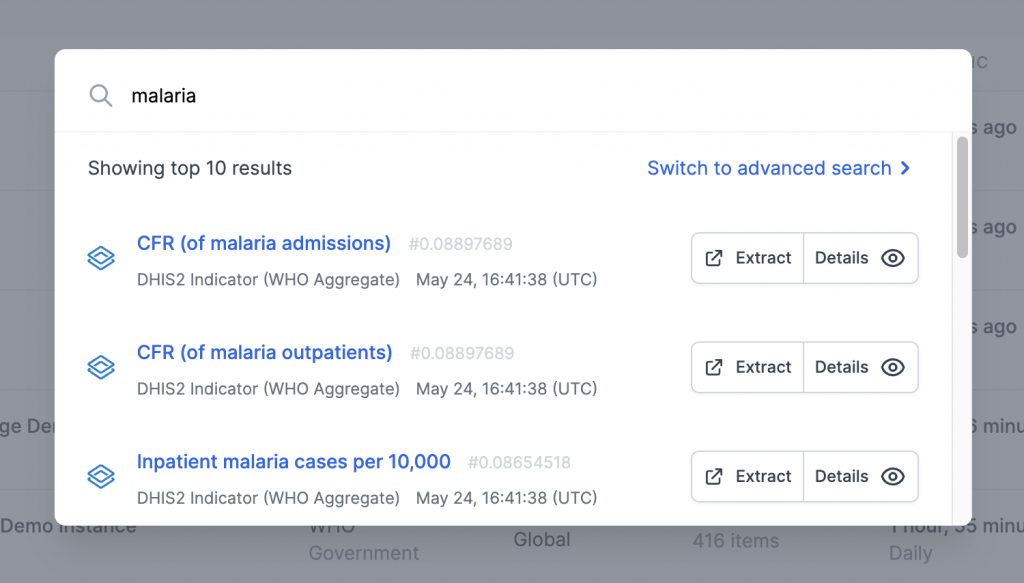
- Data exploration through the data catalog component. This allows users to browse, search and discuss data from different sources (S3 buckets, PostgreSQL databases, DHIS2 instances…). It will make possible the synchronization of data from different sources and formats.
- Collaborative interactive computing through the notebooks component, based on Jupyterhub, where users can create and share notebooks. This will allow software users to easily share their ideas and advancements across teams.
- Automated data extraction and transformation using the data pipelines component, which will greatly speed up the extraction processes.
- Data visualization through the visualization component. This provides an easy way to use OpenHexa data in different data visualization and business intelligence tools.
OpenHexa also provides powerful access control features. These allow you to make sure that users can only see and work on specific data for which they have authorization.
A user oriented open source platform
By developing OpenHexa as an open-source platform, we are tackling two challenges:
- The lack of open-source integrated data platforms: there are many high-quality open-source software tools for data visualization, analysis or automation, but integrated platforms that combine the different aspects of data science tend to be proprietary, expensive, and plagued by opaque pricing structures.
- The brittleness of data workflows in public health projects: when working with health data, experts often face heterogeneous data, tasks that require manual operations, and siloed information, leading to disorganized analyses that are and difficult to reproduce .
OpenHexa offers a novel solution that is both integrated and 100% open-source (codebase available on Github). Bluesquare can host the platform in its cloud infrastructure, or you can deploy it yourself on any cloud provider.
Our aim with developing this platform is to offer our current and future partners a tool that enables programs and managers to automate processes that are often manual, time consuming and error-prone. We also had at heart to make it open source and to have a user friendly interface.
Are you as excited about the launch of Openhexa as we are? Participate in the development of the source code on Github or request a demo session with one of our experts.
Use case: Improving the national surveillance system for infectious diseases
While not limited to health-centric workflows, our platform has been developed with health data as the primary use case.
OpenHexa offers an ideal environment for local universities, analytical units within Ministries of Health, Institutes of Public Health, National Statistical Institutes or international partners to implement a wide variety of data analysis, at national or sub-national level.
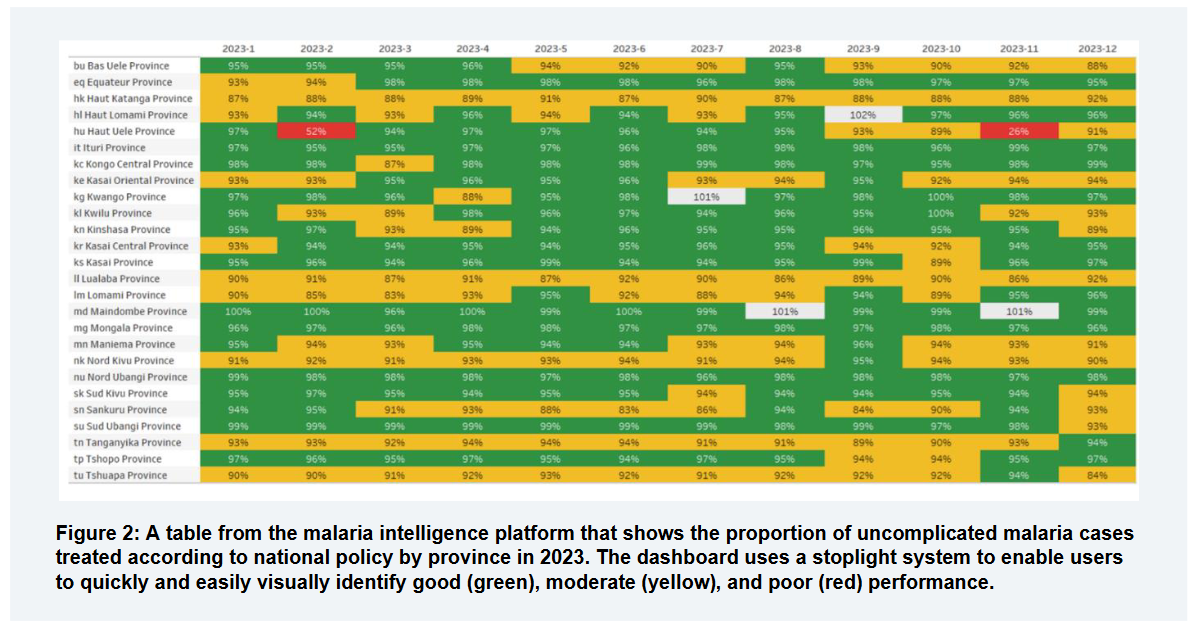
As an example, let’s consider an epidemiologist who wants to improve the national surveillance system for infectious diseases. This system relies on weekly data collection on about 20 diseases collated at the district level. Each week, a data manager manually aggregates data and sends it to the national surveillance team at the MOH. Then analysts from the national surveillance team evaluate the data using Microsoft Excel and try to identify outbreaks.
How can OpenHexa improve this workflow?
- Using the data pipelines component, automated extraction pipelines are implemented to ensure that up-to-date data is consolidated every week. Current use cases include data coming from DHIS2, Excel systems, EpiData collection systems, Access databases
- Within the notebook component, a data scientist develops an outbreak detection algorithm in collaboration with national experts and academic teams and colleagues within and outside the country
- The data scientist can share the outbreak notebook with their colleagues and local experts
- The outbreak algorithm is then deployed as a data transformation pipeline and scheduled to run every week using the latest data
- Using a third-party visualization tool (such as Tableau or PowerBI) connected to OpenHexa, a data visualization expert creates a dashboard to visualize the outbreak data
- A local monitoring and evaluation team is trained to use the data integration platform, operate the outbreak detection code, and evaluate the data visualized in the dashboard, and oversees regular updates to the surveillance system. They can zoom on specific zones, compare outbreak cinetics from different years, articulate various relevant data series.
Thanks to OpenHexa, we have moved from a manual, error-prone process to an automated and reproducible solution. This allows in-country teams to generate better insights into district-level epidemiological trends.